plyxp provides efficient abstractions to SummarizedExperiment such that using common dplyr functions feels as natural to operating on a data.frame or tibble. plyxp makes use of a concise grammar for exploring and manipulating annotated matrix data in the form of the SummarizedExperiment, scaling from simple to complex operations spanning one or more tables of data. Some of the optimized implementations in plyxp power functionality within the tidySummarizedExperiment package, which also offers a dplyr-like interface to SummarizedExperiment. These two packages can easily be used in parallel, by casting objects with the new_plyxp constructor to enable plyxp-driven functionality.
plyxp uses data-masking from the rlang package in order to connect dplyr functions to SummarizedExperiment slots in a manner that aims to be intuitive and avoiding ambiguity in outcomes.
Note: This package is still under active development. Feel free to reach out to the package developers, see Feedback section below.
installing plyxp
# plyxp is available on BiocManager version 3.22
BiocManager::install("plyxp")
# To use the latest updated version please use the github
remotes::install_github("jtlandis/plyxp")data masking SummarizedExperiment
The SummarizedExperiment object contains three main components/“contexts” that we mask, the assays(), rowData()1 and colData().
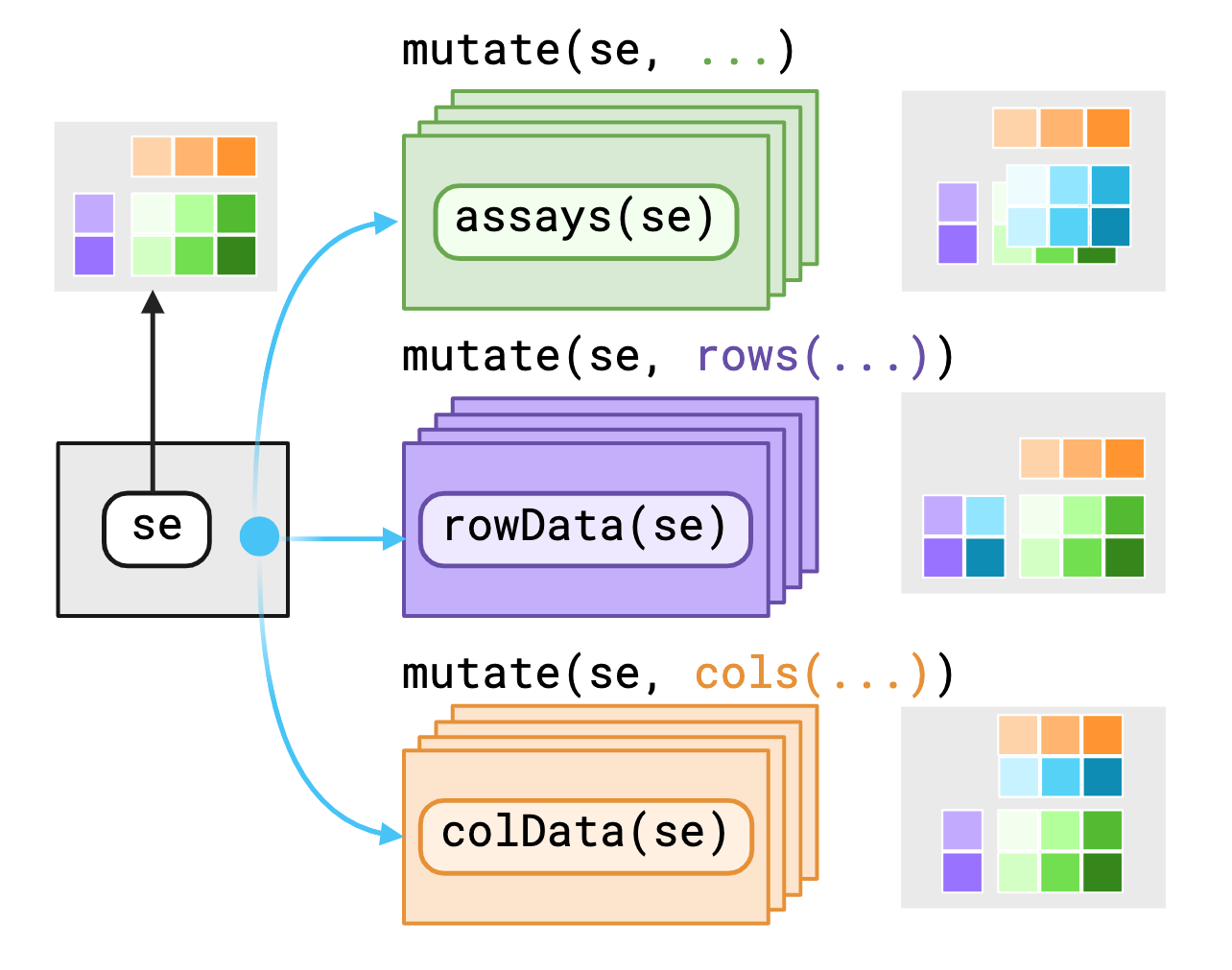
plyxp provides variables as-is to data within their current contexts enabling you to call S4 methods on S4 objects with dplyr verbs. If you require access to variables outside the context, you may use pronouns made available through plyxp to specify where to find those variables.
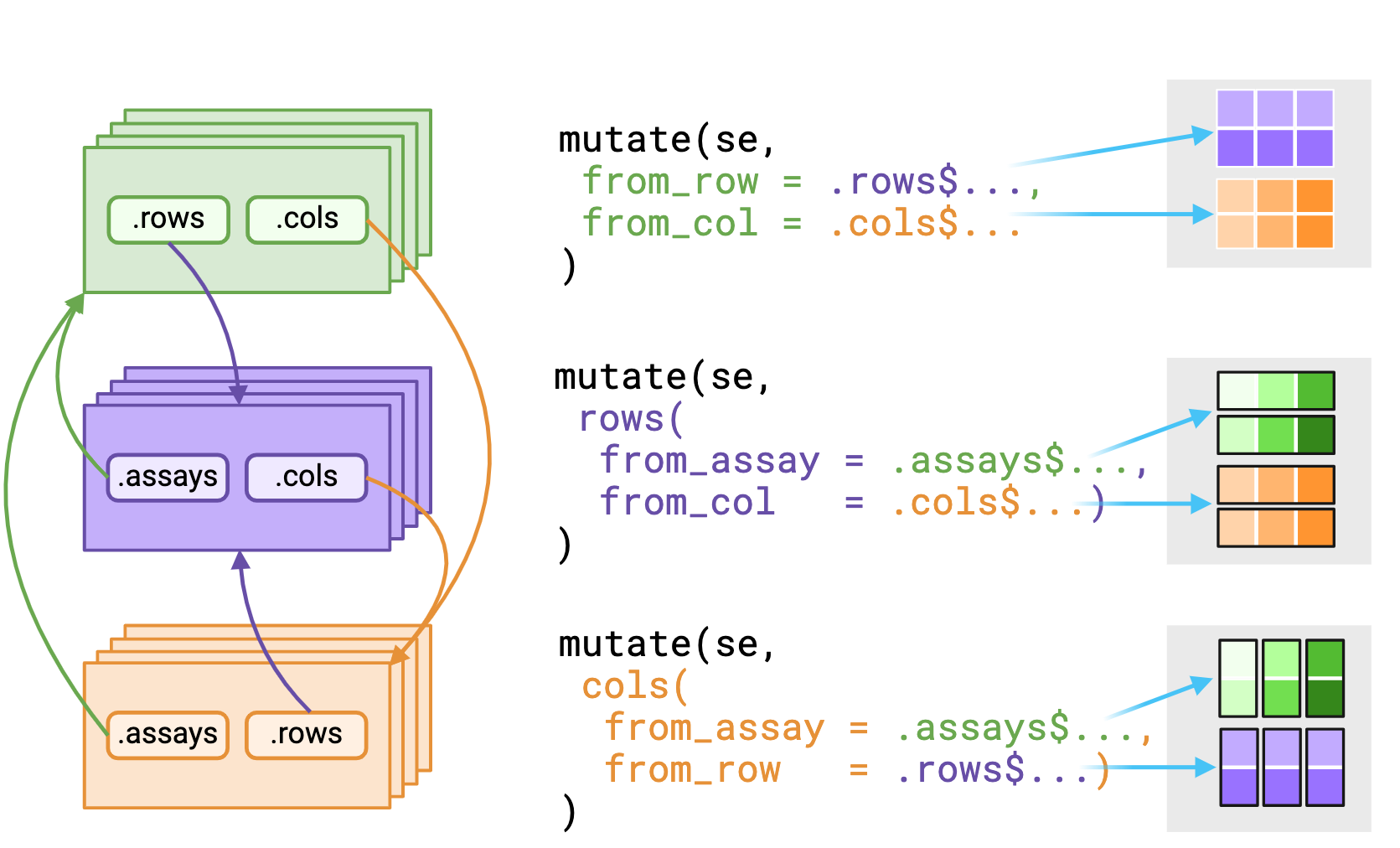
_asis variant that returns underlying data without reshaping it to fit the context. Figure made with Biorender
The .assays, .rows and .cols pronouns outputs depends on the evaluating context. Users should expect that the underlying data returned from .rows or .cols pronouns in the assays context is a vector, replicated to match size of the assay context.
Alternatively, using a pronoun in either the rows() or cols() contexts will likely return a list equal in length to either nrows(rowData()) or nrows(colData()) respectively.
Feedback
We would love to hear your feedback. Please post to Bioconductor support site or the #tidiness_in_bioc Slack channel on community-bioc for software usage help, or post an Issue on GitHub, for software development questions.
Note to Users
plyxp is still under active development. We have recently discovered an error in group_by(xp, rows(foo)) |> summarize(some_assay = <expr>) operations in which the resulting assay matrix was being collected incorrectly. This has been fixed with this commit and has been pushed to plyxp 1.4.3 on Bioconductor version 3.22. With this being said, we cannot update older version of plyxp on Bioconductor 3.21 and 3.20 - however we have cherry-picked this commit into the github branch images.
Thus if you wish to use plyxp from Bioconductor 3.21 or 3.20, please install from github to ensure you have the latest fixes.
remotes::install_github("jtlandis/plyxp@RELEASE_3_21")